What are Biochemical Pathways?
Biochemical pathways are the sequences of chemical reactions that occur in living cells, such as metabolism, signaling, and gene expression. They are essential for understanding the molecular mechanisms of human biology and disease, as well as for developing new drugs, therapies, and understanding their interactions. Conveniently, there are several online resources that provide comprehensive and curated collections of biochemical pathways, as well as tools for exploring and analyzing them. Three of the most popular and widely used resources are Reactome, Kyoto Encyclopedia of Genes and Genomes (KEGG), and WikiPathways.
Reactome
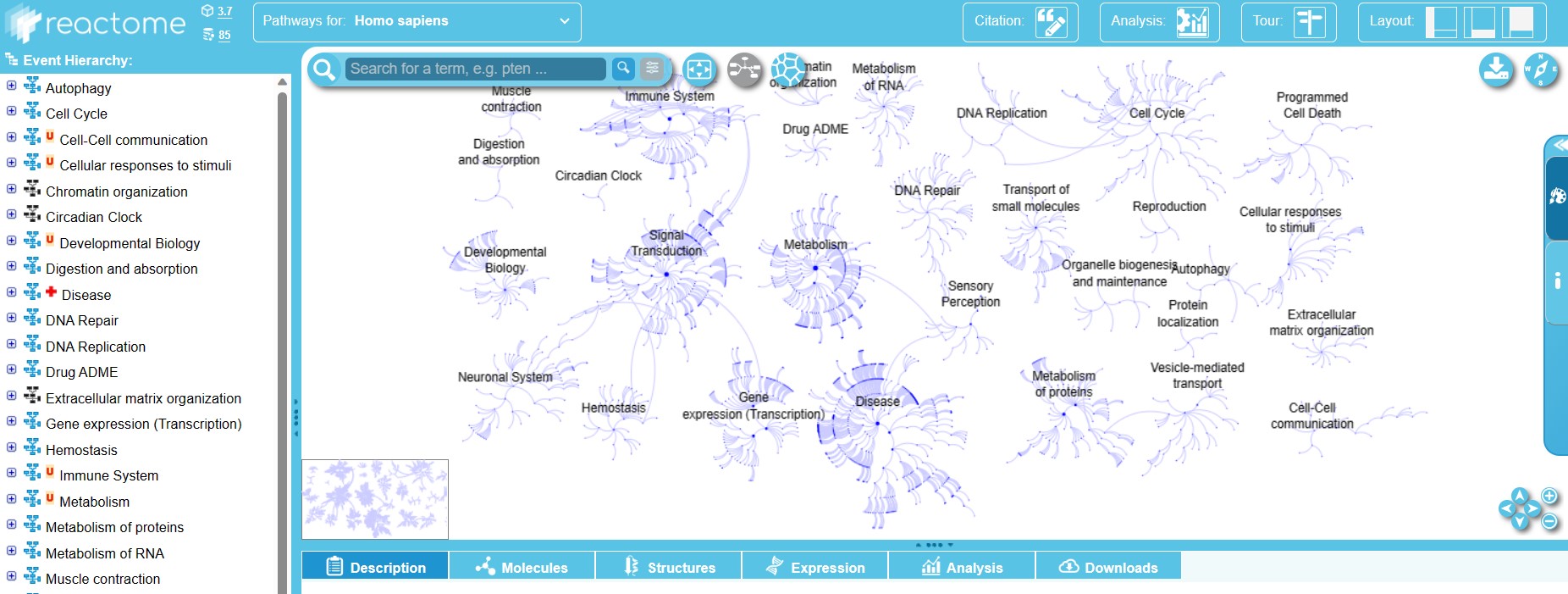
Reactome is a free, open-source, and community-driven database of human biochemical pathways. It covers various aspects of human biology, such as cellular processes, physiological systems, diseases, and drug targets. Reactome provides a hierarchical organization of pathways, from simple reactions to complex events. It also provides detailed annotations of pathway components, such as molecules, interactions, locations, and functions. It also offers interactive pathway diagrams that show the structure and dynamics of pathways, as well as links to external databases and literature. Reactome also provides tools for pathway analysis, such as enrichment analysis, expression analysis, and network analysis. These tools allow users to identify relevant pathways for their own data sets, such as gene lists or expression profiles.
Kyoto Encyclopedia of Genes and Genomes (KEGG)
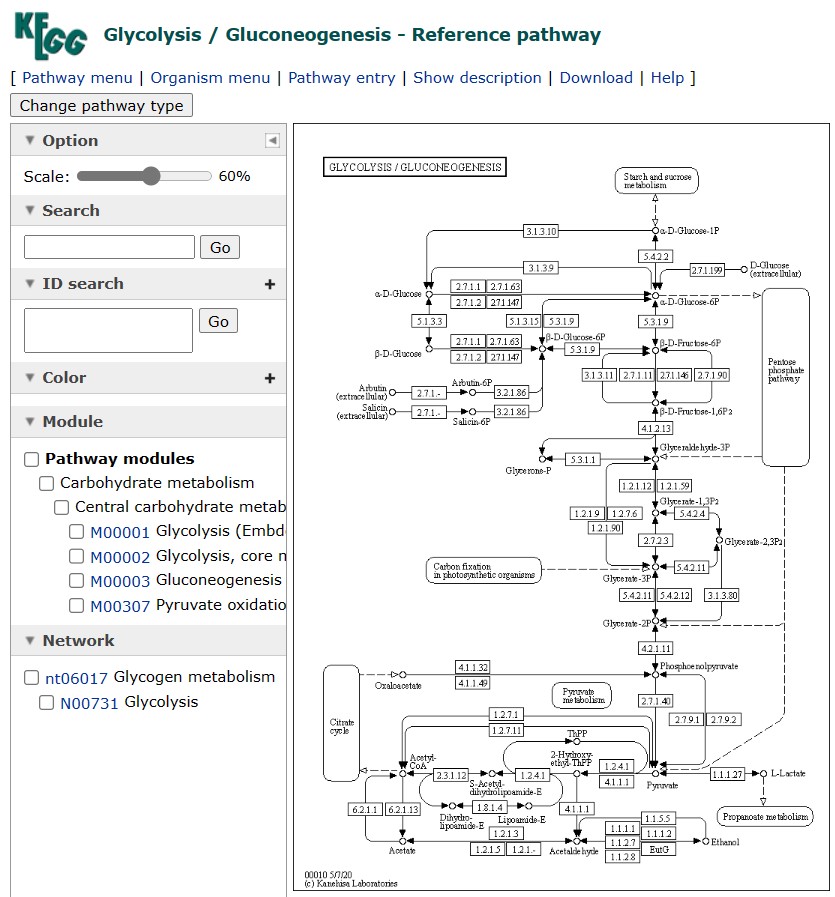
KEGG is a comprehensive database of biological systems, including biochemical pathways, genomes, diseases, drugs, and chemical substances. It covers various domains of life, such as bacteria, archaea, eukarya, and viruses. KEGG provides a systematic representation of pathways based on the concept of modules, which are functional units of genes or molecules that perform a specific biological task. KEGG also provides annotations of pathway components, such as genes, proteins, metabolites, reactions, and enzymes. It also offers graphical pathway maps that show the relationships and interactions among pathway components, as well as links to external databases and literature. KEGG also provides tools for pathway analysis, such as pathway mapping, comparison, and integration. These tools even allow users to map their own datasets onto KEGG pathways or compare them with other datasets.
WikiPathways
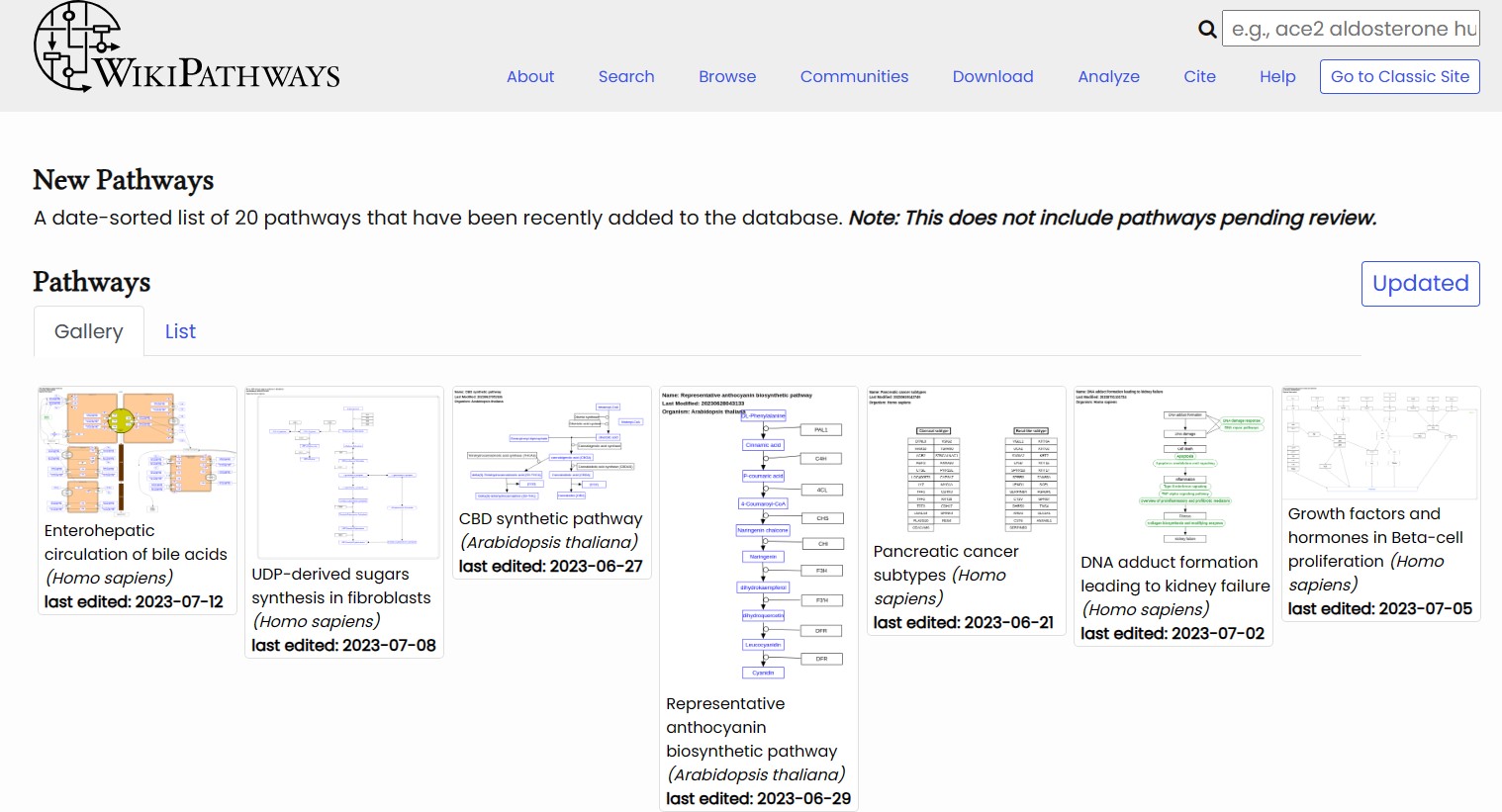
WikiPathways is a collaborative platform for creating, editing, and sharing biochemical pathways. It covers various species and topics of biology, such as human health and disease, plant biology, animal models, and environmental toxicology. WikiPathways provides a wiki-based approach for pathway curation and annotation, where anyone can contribute to the content and quality of pathways. WikiPathways also provides graphical pathway diagrams that show the layout and information of pathway components using standard notations and symbols. It also offers interactive pathway viewers that allow users to explore and manipulate pathway diagrams online or offline. WikiPathways also provides tools for pathway analysis, such as enrichment analysis and network analysis. These tools allow users to find relevant pathways for their own datasets or build networks from pathway components.
Sources
WikiPathways(2023). https://www.wikipathways.org/.
KEGG PATHWAY Database (2023). Kyoto Encyclopedia of Genes and Genomes (KEGG). https://www.genome.jp/kegg/pathway.html.
Reactome (2023). https://reactome.org/PathwayBrowser/.
